The Agar Root Phenotyping System represents a groundbreaking solution that redefines approaches to root research. Driven by state-of-the-art robotic technology, the system effortlesly handles PSI-developed Petri plates around the clock, throughout the every step of the phenotyping process. This encompasses tasks ranging from agar pouring and robotic seed sowing to the precise application of bacteria at the root-stem interface, and to comprehensive phenotypic evaluation conducted at various imaging stations.
Reduce lab work, increase scientific exploration
- Fully automated root & shoot screening system
- Automation through robotic manipulation
- Plants grown in vitro on agar plates
- Comprehensive end-to-end workflow solution
- Automated media pouring
- Precise robotic seeding and seed sorting
- Exact application of liquid solutions onto the specific root site
- High-content imaging of multiple fluorescence proteins
- High-resolution imaging of chlorophyll fluorescence kinetics
- Absorption and fluorescence hyperspectral imaging
- Cutting-edge neural network-based root image analysis
Agar Root Phenotyping System
The Agar Root Phenotyping System is designed for high-throughput, automated, software-controlled phenotyping of plants grown in vitro. The system is customized for efficient operation with PSI developed square Petri plates, and can accommodate thousands of plates at full capacity.
The Agar Root Phenotyping System is engineered to offer a comprehensive end-to-end workflow solution requiring no human intervention. The process initiates with sample preparation and cultivation, encompassing tasks such as media pouring and automated robotic seeding. It progresses through stratification at 5 °C to long-term cultivation of samples in “Growth Hotels”. Complete automation through robotic manipulation facilitates the integration of multiple imaging stations. These enable continuous non-invasive monitoring of root morphology, the detection of expression of various fluorescent proteins in plant roots, and the measurement of chlorophyll fluorescence kinetics in plant shoots. Uniquely, the system also includes fluorescence hyperspectral imaging of coumarins production. Additionally, it incorporates an application station that enables the sterile application of bacteria to a specified root site. The entire system is could be designed with an emphasis on sterility, and manipulation with GMO plants and bacteria.
Effortless Plate Preparation
A crucial component of the Hades system is the Plate Preparation Table. This remarkable engineering achievement provides seamless, fully automated sample preparation in a sterile atmosphere established through HEPA-based air filtration and UV-C sterilization. The table is capable of accommodating up to 1500 sterile Petri plates.
The cutting-edge platform is equipped with an automated agar pouring mechanism, guaranteeing precise filling of media from a commercial media preparation device. The station relies on two SCARA robots and conveyor belts to manage all aspects of sterile plate handling, eliminating the need for human intervention. Additionally, the platform includes a seeding robot, Boxeed, which sows seeds onto the agar in precise accordance with predefined parameters. The process involves image analysis of each seed to identify and select viable individuals.
Optimized Growth Environments
The Growth Hotel is a shelving cultivator designed exclusively for cultivating plant material on Petri plates. Within each Hotel, there are 4 shelves, each accommodating transport cassettes that each hold 20 plates. With the ability to hold 9 cassettes on each shelf, the total capacity of each Growth Hotel module is an impressive 720 Petri plates.
The Growth Hotel is equipped with multichannel LED illumination. Irradiance (up to 500 µmol.m-2.s-1), spectral quality and specific light regimes are adjustable for each shelf and each color channel separately via a user-friendly interface.
A dedicated section within the growing space is responsible for controlling seed stratification at a precisely maintained temperature of 5°C.
Cutting-Edge Imaging Insights
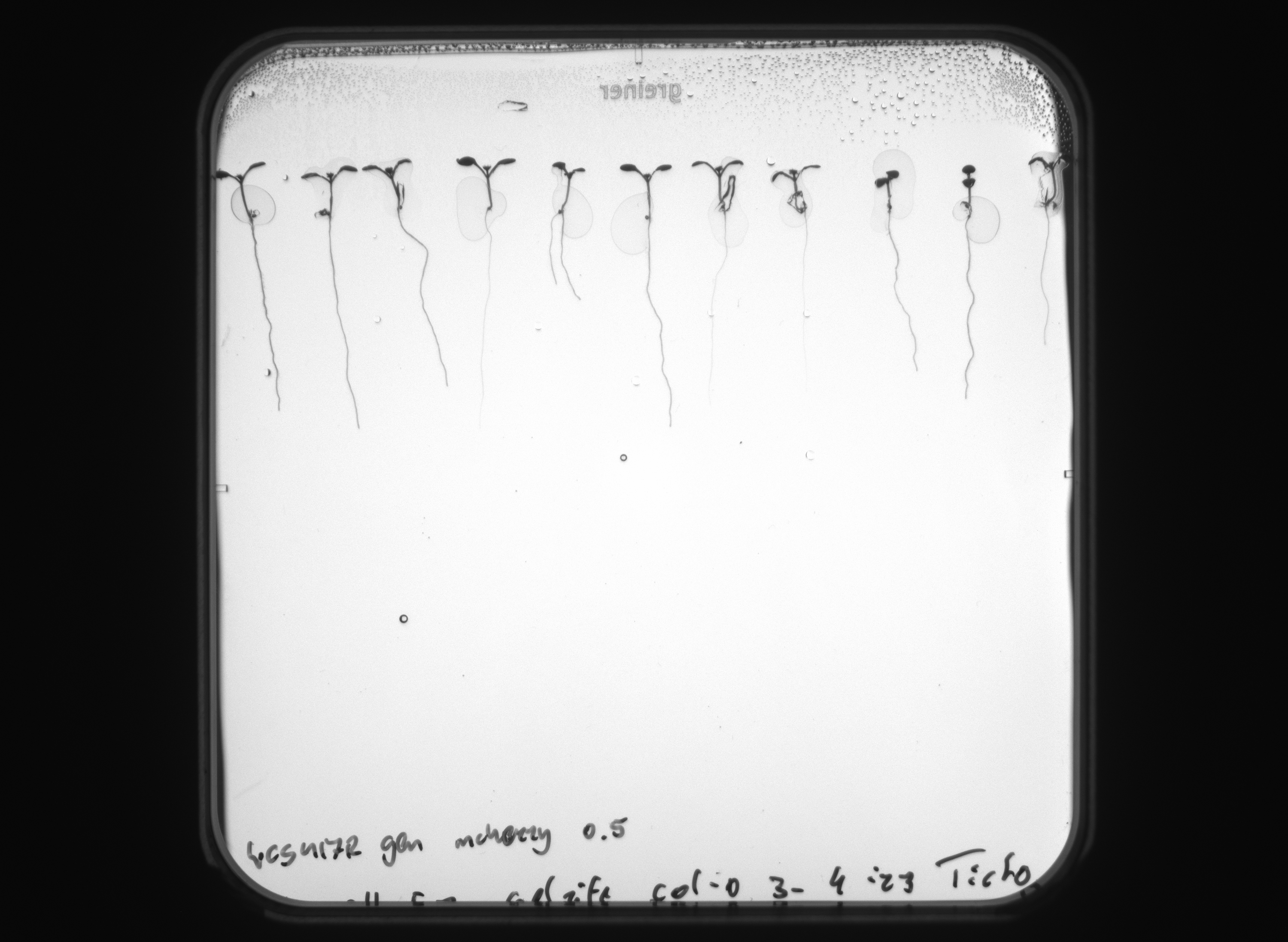
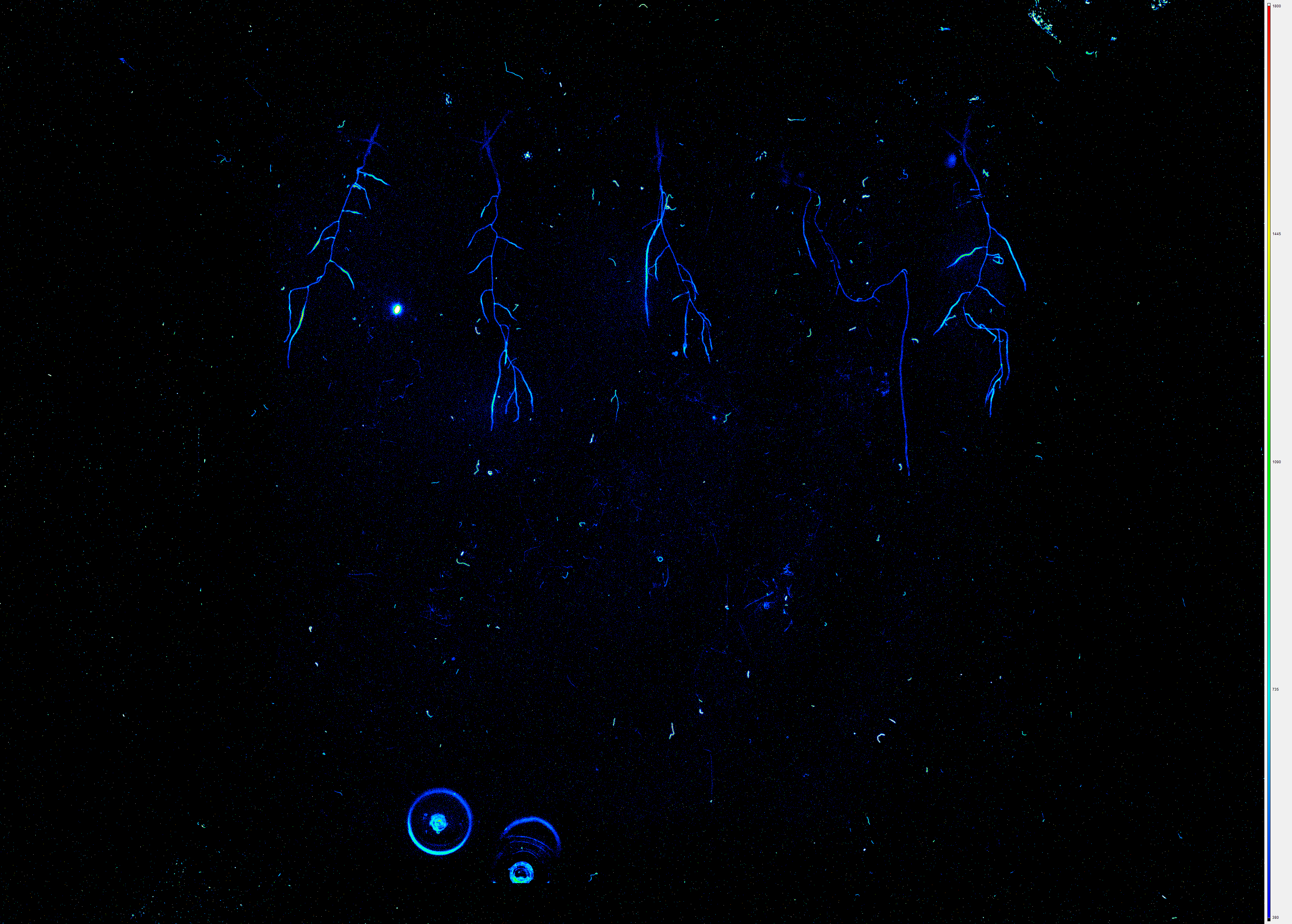
Guided by robotic arms, the samples are moved from the cultivation hotels to imaging stations for the study of root and shoot development. These stations capture chlorophyll fluorescence data, and compounds such as fluorescent proteins (GFP, YFP, RFP, mCherry) and coumarins are visualized without sample disruption. Hyperspectral cameras provide an extra layer of imaging sophistication, revealing the biochemical status of samples.
The state-of-the-art AI technology is utilized to analyze traits from the acquired images. The software employs one of the most renowned computer vision methods, Convolution Neural Networks (CNNs), to effectively segment root objects from the background.
Software
Comprehensive software package has been developed for system control, data acquisition, image analysis and database configuration. User friendly graphical interface is designed to control all hardware system components actions, to control and monitor environmental conditions and to design experiments with an extremely high level of flexibility.
For easier and user friendly control, system is equipped with Human Machine Interface with touch-screen display. Scheduling assistant with calendar function allows running multiple experiments simultaneously, provides different modes for experiment randomization, for treatment per plant or group of plants with different experimental protocols and plant handling. Software also allows users to set amount of media poured into plate and adjust seeding and pipetting protocols.
All acquired imaging, sedding and manipulation data are stored in an SQL database, processed and available for inspection and further analysis in range of seconds after recording via user-friendly graphical interface. PlantScreenTM Data Analyser provides tools for data browsing, grouping, analysis, user-defined reprocessing and export. Multiple clients can be connected to the database, with different privileges assigned based on a built-in authentication mechanism. An SMS and mail notification service is integral part of the complete phenotyping system. 24-hour online support service is key component of the PlantScreenTM phenotyping solution.
Key features of IT solution
- Comprehensive software package for system control, data acquisition, image analysis and data base configuration
- Open database structure
- Flexibility to integrate custom external analyses
- Data accessibility via the open Rest API
- Root & shoot analysis based on Convolution Neural Networks (CNNs)
- Remote access
- Automatic SMS and email notification service
- 24-hours online support service
Why to use PlantScreenTM Phenotyping System
In the search for beneficial traits that may allow crops to resist abiotic and biotic stresses, fast and accurate methods are required for efficient and effective plant high-throughput phenotyping. Such methods must involve automated measurements of plant morphology, biochemistry and physiology to determine potential and actual yield under a variety of monitored environmental conditions. With over 20 years’ experience of designing instrumentation for plant imaging PSI is now at the forefront of providing complete solutions PlantScreenTM Phenotyping Systems for automated multidimensional plant phenotyping.
PlantScreenTM Phenotyping Systems designed for integrative phenotyping on temporal and spatial level are proven in a range of applications across the world:
- High-throughput screening
- Morphology and growth assessment
- Nutrient management
- Photosynthetic performance
- Abiotic stress
- Pathogen interaction
- Trait identification
- Chemical screening
- Nutrient effects
Customized solution
The Agar Root Phenotyping System is modular and designed so that additional features may be added as the user´s screening requirements evolve. Further standard layout in terms of dimensions, capacity and range of image-based sensors can be adapted in order to meet your specific research question.
Furthermore we make our state-of–art phenotyping facilities available for testing and proof-of-concept experiments. Contact us for more information.
Discuss your project and test your concept
The nature of the research questions is unique referring to distinct requirements you might have to answer your specific questions. Our goal is to consult with you extensively your project to ensure that our technology is configured to meet your needs precisely. At PSI we have team of plant physiologists, image-processing specialist and technical engineers that can help to shape your phenotyping projects to meet precisely your requirements.
FluorCam
The custom-designed High-Sensitivity Kinetic Chlorophyll Fluorescence Imaging Unit & Morphometric Unit is based on Flat FluorCam FC1300/2020 with Advanced Multi-Excitation Module and integrated 7-position motorized filter wheel, tailored for imaging of weak fluorescence signals from Fluorescence Proteins as well as Chlorophyll fluorescence kinetics and it also enables analysis of roots morphology in transmission mode. The system is optimized for roots imaging on square Petri dishes. The unit is fitted with the 12.36-megapixel CMOS sensor (Sony IMX253LQR-Q). The sensor delivers a resolution of 4112 × 3006 pixels as well as global shutter feature. The sensor is extremely sensitive and produces sharp, low-noise images.
| Monochromatic Sensor | |
|---|---|
| Sensor Technology | CMOS Monochromatic |
| Resolution (MPix) | 12.36 (3.09 in binning mode) |
| Resolution (H × W) | 4112 × 3006 (2056 × 1503 in binning mode) |
| Bit depth | 12 bit |
| Sensor Size | 1.1" |
| Shutter | Global Shutter |
| Max. fps in freerun mode | 2 |
| Sensor Model | Sony IMX253LLR-Q |
| Pixel size | 3.45 µm (6.9 µm, in binning mode) |
| Design | |
|---|---|
| Interface | GigE |
| Lens Mount | F - Mount |
| Camera Dimensions (H × W × L) | 200 mm × 132 mm × 95 mm |
| Power supply | 12 V - 24V |
| Lights | |
|---|---|
| Chlorophyll fluorescence module | Red-Orange (620 nm), Cool-White (5700K), Far-Red (735nm) |
| Multicolor module | UV (365 nm), Royal Blue (450 nm), Blue (470 nm), Cyan (505 nm), Green (530 nm), Amber (590 nm) |
| Morphometric module | Cool-White (5700K) |
| Emission filters | |
|---|---|
| Filters | F469, F483, F513, F565, F586, F593, F520, F635, glass, |
VNIR Hyperspectral Imaging Unit
The VNIR Hyperspectral Imaging Unit was specially developed for measurement of fluoresce substances in the root system of plants grown on square Petri dishes. The UV light of 365nm is used for excitation of fluorescent emission signals in the visible and near infrared region of the spectrum (380 – 900 nm). Step-scanning mode enables the use of long integration times when imaging the fluorescence signals. The unit also contains white light which is used in a transmission geometry to generate a plant and root outline mask.
| Imaging & Optical Data | |
|---|---|
| Spectral Range | 350 – 900 nm |
| Band Size | 520 nm |
| Entrance Slit Width | 50 µm |
| Dispersion/Pixel | 0.28 nm/pixel |
| Wavelength Resolution, FWHM | 2 nm |
| Spatial Resolution | 500 pixels |
| Spectral Resolution | 480 pixels |
| Image Frequency | 45 fps |
| Sensor | |
|---|---|
| Type | CMOS |
| Size | 1/1.2" |
| Resolution | 1,920 × 1,000 pixels |
| Bit Depth | 12 bit |
| Pixel Size | 5.86 µm |
| Dynamic Range | 67 dB |
| Lights | |
|---|---|
| Reflectance mode | White |
| Fluorescence mode | UV |
| Connection | |
|---|---|
| Control & Data | GigE |
































.png)
